近期,中南大学湘雅医院神经外科刘庆教授团队在国际知名期刊《Exploration》(IF=22.5)上发表了题为“Deciphering the Potential Causal and Prognostic Relationships Between Gut Microbiota and Brain Tumors: Insights from Genetics Analysis and Machine Learning”的研究论文。该研究基于大规模遗传数据与多组学整合分析,首次系统揭示了肠道菌群与三种常见脑肿瘤(脑膜瘤、垂体瘤和胶质瘤)之间的潜在因果关系,并成功构建了基于微生物相关基因(MRGs)的胶质瘤预后预测模型(MRS),为脑肿瘤的早期风险预警、预后评估及免疫治疗提供了新思路。湘雅医院神经外科吴长武副教授为论文第一作者,刘庆教授、谭军助理研究员为论文通讯作者。
https://doi.org/10.1002/EXP.20240087
脑肿瘤是中枢神经系统常见疾病,其中恶性胶质瘤预后极差,目前尚缺乏有效的早期诊断与预后预测标志物。近年来,“菌群—肠—脑轴”调控机制备受关注,多项观察性研究提示脑肿瘤患者存在肠道菌群紊乱,但其因果关联及临床意义仍未明确。
本研究依托大型基因组数据库(包括MiBiOGen、FinnGen、TCGA等),运用多种孟德尔随机化(MR)及机器学习方法,首次从遗传角度探索了肠道菌群与脑肿瘤的潜在关联(图1)。首先,本研究证实特定肠道菌群对脑肿瘤的因果效应:识别出4种与脑膜瘤相关的菌群、7种与垂体瘤相关的菌群,以及8种与胶质瘤相关的菌群(图2)。考虑到肠道微生物的丰度可能相互影响,本研究对可能与三种脑肿瘤存在因果关系的肠道微生物进行了MVMR‐IVW分析,以探讨不同微生物在因果关系中的重要性。对于脑膜瘤,粪球菌属(Coprococcus 1)在调整其他微生物后因果关系不再显著,表明其效应可能受其他菌群影响;垂体瘤中所有七种微生物仍显著,提示其独立作用;而在胶质瘤中,仅梭菌纲(Clostridia)保持显著,说明其可能起主导因果作用(表1)。鉴于肠脑轴的双向通讯,肠道菌群的丰度可能会受到脑肿瘤的影响。我们根据脑肿瘤的GWAS数据进行了反向双样本MR分析,没有证据表明这三种脑肿瘤可能对肠道菌群的丰度产生因果效应。
Figure 1. Study design and workflow. The blue blocks represent data sources or analyses that contain data sources. The yellow blocks represent the selection of IVs, and the gray blocks represent the conducted analysis.
Figure 2. Two sample MR results of causal effects of gut microbes on meningioma (A), pituitary tumor (B), and glioma (C). Data are expressed as an odds ratio (OR) with corresponding 95% confidence interval (CI). The forest plots also include the results of sensitivity analyses. *p < 0.05, **p < 0.01.
进一步,研究团队创新性地利用Sherlock Bayesian统计框架识别脑中影响特定肠道微生物丰度的微生物相关基因(Microbiota-Related Genes,MRGs)。该分析整合了大脑皮质eQTL数据与GWAS摘要数据,成功鉴定出在脑组织中表达的103个与脑膜瘤相关的MRGs、40个垂体瘤相关MRGs及63个胶质瘤相关MRGs。为了进一步探索MRGs在脑肿瘤中的潜在临床意义,研究团队收集了多个脑肿瘤转录组队列。在脑膜瘤队列中发现,与脑膜瘤有因果关系的每种微生物相对应的MRG都具有与脑膜瘤WHO分级相关的特定基因(图3A-D)。在预后价值方面,研究团队发现多个脑膜瘤相关的MRGs与脑膜瘤的无复发生存期密切相关(图3E),表明了这些MRG作为预后生物标志物的可行性。在垂体瘤队列中,本研究总共确定了五个与垂体瘤侵袭性相关的MRGs(MED27、RARRES2、AKAP8L、RAB5B和STK39)(图3F-L)。
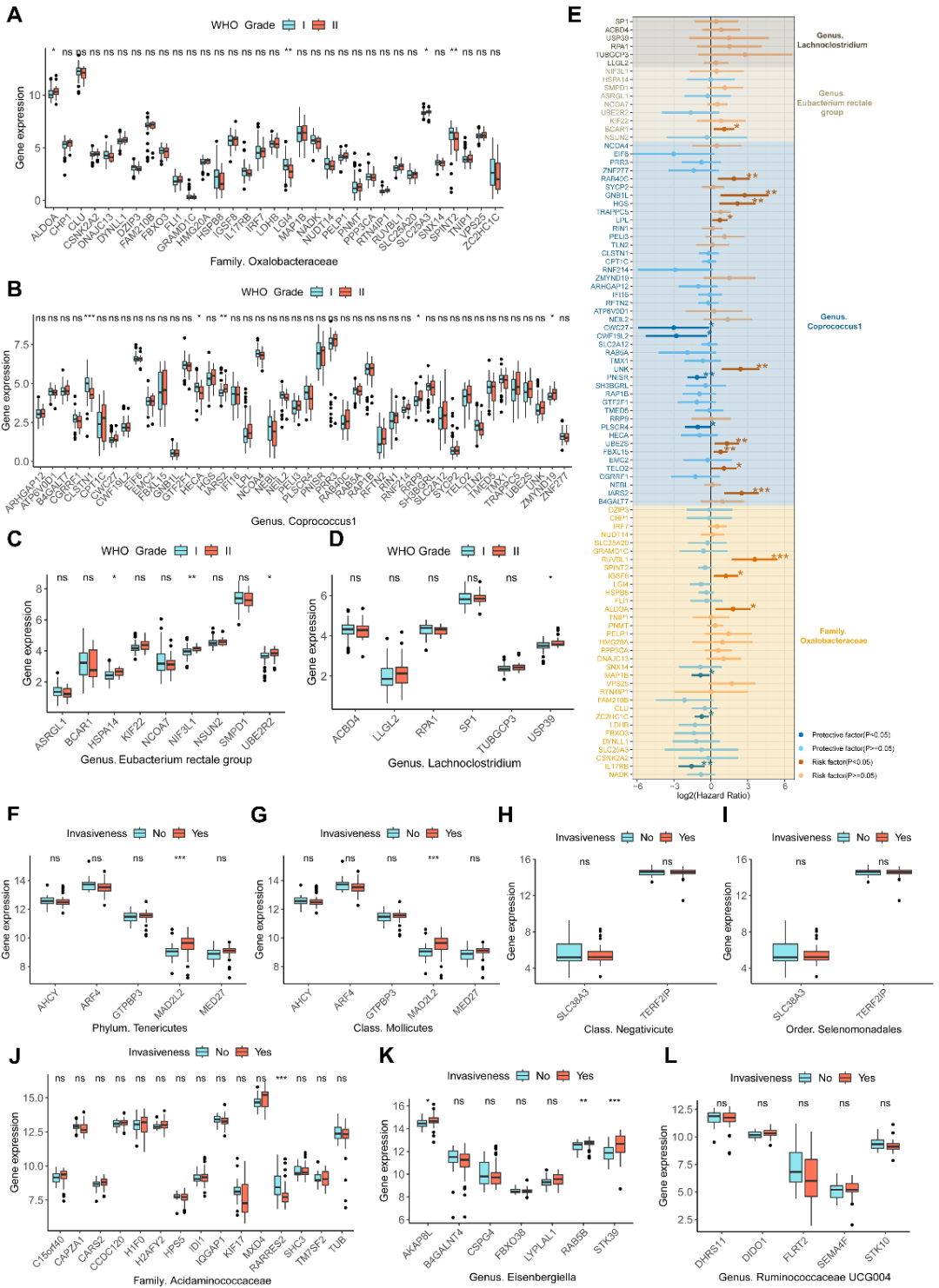
Figure 3. The relationship between MRGs in meningiomas and pituitary tumors and clinical characteristics. (A–D) The expression differences of meningioma‐related MRGs between WHO grade I and II meningiomas. Figures (A–D) respectively represent the expression of MRGs corresponding to the gut microbe that has a causal effect on meningiomas. (E) The forest plot displays the results of univariate Cox regression analyses of meningioma‐related MRGs on the recurrence‐free survival of meningiomas. The different colored squares correspond to MRGs of different gut microbes. (F–L) The expression differences of pituitary tumor‐related MRGs between invasive and non‐invasive pituitary tumors. Figures (F–L), respectively represent the expression of MRGs corresponding to the gut microbe that have a causal effect on pituitary tumors. *p<0.05, **p<0.01, ***p<0.001.
对于胶质瘤,研究团队首先使用TCGA队列探讨了MRGs与胶质瘤临床特征之间的关联。除SOX15外,所有MRGs均与胶质瘤的WHO分级显著相关(图4A-H)。此外,几乎所有MRGs都与胶质瘤的总生存期(OS)密切相关(图4I),这表明MRGs在胶质瘤的发展和预后中具有潜在意义。由于MRGs的表达可作为肠道微生物丰度的间接指标,这也意味着肠道微生物群的丰度可能影响胶质瘤的预后。
Figure 4. The relationship between MRGs in gliomas and clinical characteristics. (A–H) The expression differences of glioma‐related MRGs between different WHO grades. Figures (A–H) respectively represent the expression of MRGs corresponding to the gut microbe that have a causal effect on gliomas. (I) The forest plot displays the results of univariate Cox regression analyses of glioma‐related MRGs on the OS of gliomas. The different colored squares correspond to MRGs of different gut microbes. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
基于上述发现,研究团队整合了来自TCGA、CGGA、Rembrandt共1688例胶质瘤样本的转录组数据,采用涵盖CoxBoost、plsRcox、随机生存森林等10类机器学习算法的101种组合策略,以Leave-one-out交叉验证框架系统筛选最优预测模型。最终,CoxBoost与plsRcox联合算法表现出最佳预测效能,平均C-index达0.733(图5A)。该模型筛选出14个关键MRGs,构建出微生物相关特征标签(Microbe-Related Signature,MRS),并依据其风险评分将患者划分为高危和低危群体(图5B-F)。模型评估表明MRS在多个独立队列中具有准确且稳健的预测能力(图5G-J)。多因素Cox分析显示,在调整年龄、性别、组织学特征和IDH状态后,MRS在多个队列中依然是胶质瘤的独立危险因素。
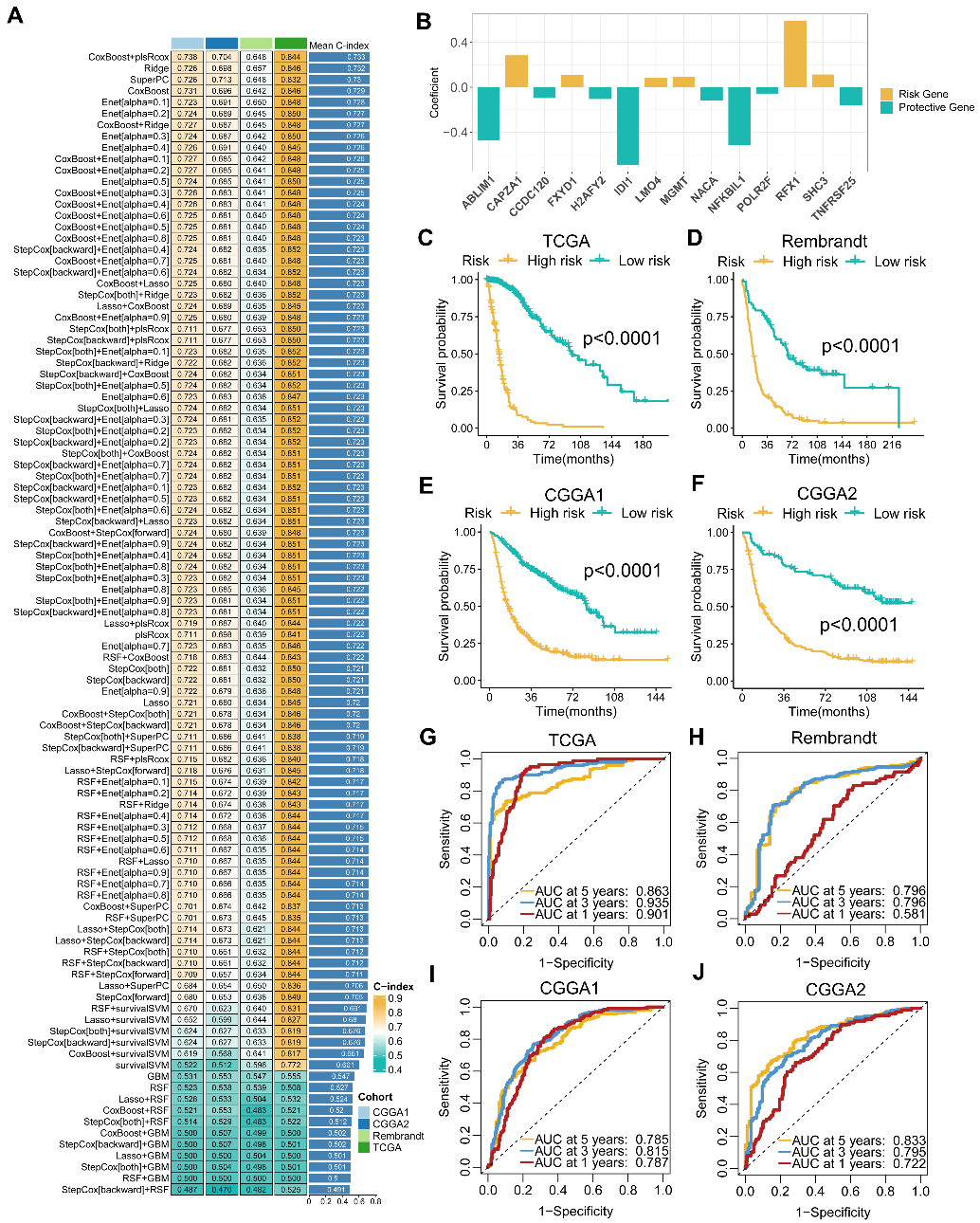
Figure 5. The establishment and validation of the MRS model via the machine learning‐based integrative procedure. (A) The 101 algorithm combination models of the integrated framework of machine learning and the C‐index of each model on all cohorts. (B) Coefficients of multivariable Cox regression analysis of 14 key MRGs finally obtained in CoxBoost+plsRcox model. (C–F) Kaplan–Meier curves depict the OS difference of glioma between MRS‐high and MRS‐low groups in TCGA (C), Rembrandt (D), CGGA1 (E), and CGGA2 (F) cohorts. Yellow represents the MRS‐high group, and green represents the pathway MRS‐low group. (G–J) ROC curves showing the OS prediction efficiency of MRS in TCGA (G), Rembrandt (H), CGGA1 (I), and CGGA2 (J) glioma cohorts.
鉴于MRS在公共队列中的可靠性能,我们进一步对来自临床内部队列(CSUXY)的151个胶质瘤样本进行了RNA测序分析,以促进MRS模型的临床转化和应用。如图6A所示,KM曲线表明内部队列中高危组的OS仍然明显差于低危组。ROC曲线显示内部队列中1年、3年和5年OS的AUC分别为0.779、0.804和0.867(图6B),这表明MRS具有出色的预测性能。此外,我们进行了多变量Cox回归显示MRS模型是内部队列的独立危险因素(图6C)。分层分析证实,MRS在GBM和非GBM队列中均具有预后价值(图6D、E)。此外,与先前已发表的100个胶质瘤预后模型相比,MRS的性能具有显著的优越性(图6F)。
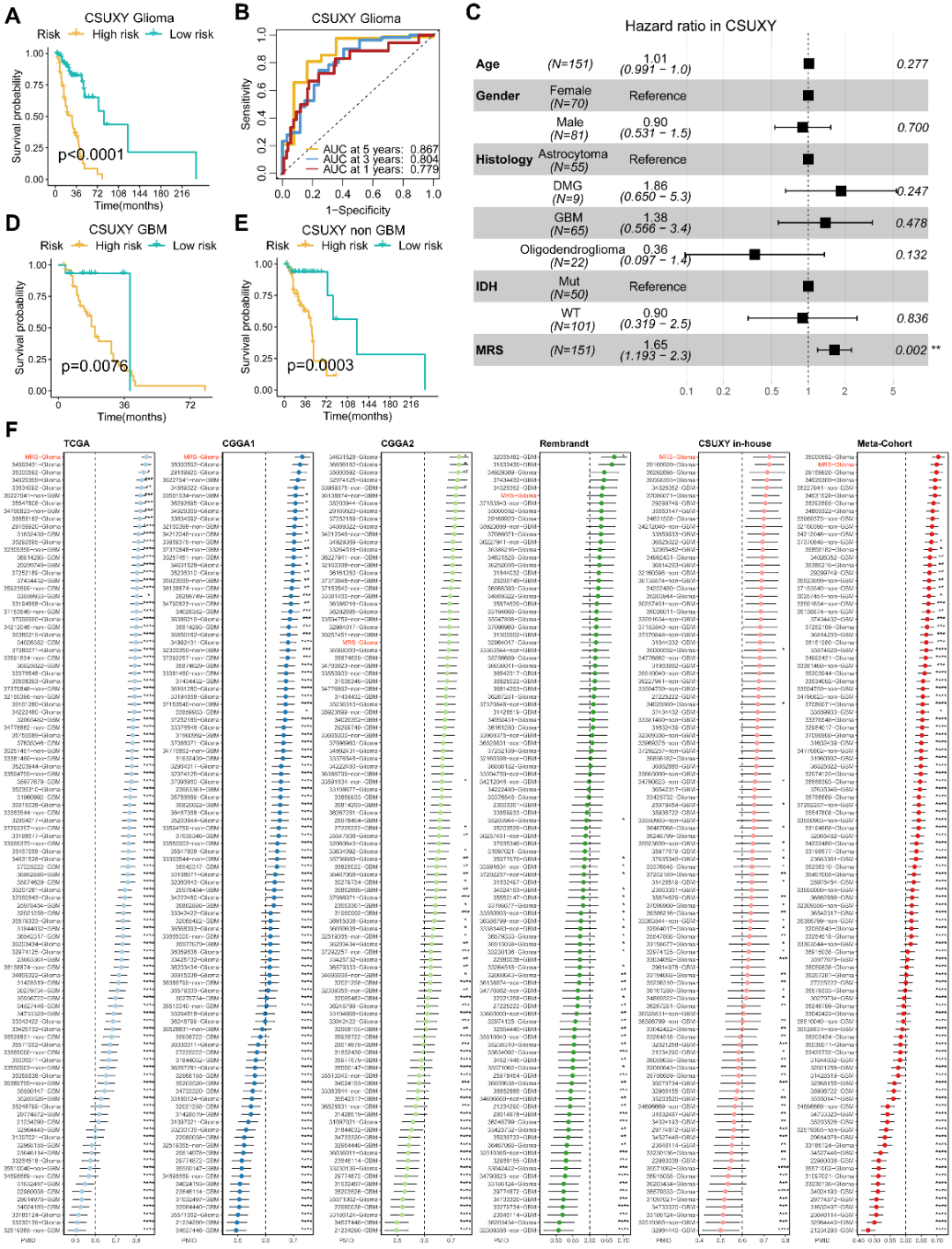
Figure 6. Validation of MRS in the clinical in‐house cohort and comparison of prognostic signatures. (A) Kaplan–Meier curve depicts the OS difference of glioma between MRS‐high and MRS‐low groups in the CSUXY in‐house cohort. (B) ROC curve showing the OS prediction efficiency of MRS in the CSUXY in‐house glioma cohort. (C) Multivariable Cox regression analysis of OS in the CSUXY in‐house cohort. Statistic test: two‐sided Wald test. Data are presented as hazard ratio (HR) ± 95% confidence interval (CI). (D) Kaplan–Meier curve depicts the OS difference of GBM between MRS‐high and MRS‐low groups in the CSUXY in‐house cohort. (E) Kaplan–Meier curve depicts the OS difference of non‐GBM between MRS‐high and MRS‐low groups in the CSUXY in‐house cohort. (F) C‐index analysis MRS and 100 published signatures in TCGA, CGGA1, and CGGA2, Rembrandt, CSUXY in‐house, and meta‐cohort. Statistic tests: Two‐sided z‐score test. Data are presented as mean ± 95% confidence interval (CI). *p < 0.05; **p < 0.01; ***p<0.001; ****p<0.0001.
团队进一步分析显示,MRS高分患者具有更显著的免疫细胞浸润特征,包括CD8+ T细胞活化与肿瘤相关巨噬细胞增多,提示其肿瘤微环境更倾向于“热肿瘤”表型(图7A-C)。最关键的是,在一个接受抗PD-1治疗的胶质母细胞瘤队列中,MRS高分患者显示更长的无进展生存期(PFS)和总生存期(OS),表明MRS不仅具备预后预测价值,还有潜力作为免疫治疗响应预测的生物标志物(图7D-G)。
Figure 7. The association between MRS and immune infiltration as well as immunotherapy response in glioma. (A–C) Heatmaps of the relationship between MRS and 28 immune cells in glioma (A), GBM (B), and non‐GBM (C) of the CSUXY in‐house cohort. Age, gender, OS status, OS time, grade, histology, and IDH status are shown as patient annotations. (D) Heatmap of the relationship between MRS and 28 immune cells in the anti‐PD‐1 cohort. (E) Correlation between MRS and the expression of PD‐1 in the anti‐PD‐1 cohort. Statistic test: Pearson's correlation coefficient. (F, G) Kaplan–Meier curves depict the overall survival (F) and progression‐free survival (G) differences of GBM between MRS‐high and MRS‐low groups in the anti‐PD‐1 cohort.
副教授,医学博士,硕士研究生导师
临床工作集中于神经肿瘤的诊治;科研工作集中于脑肿瘤的免疫微环境、肿瘤代谢、治疗抵抗以及多组学分析,旨在探索脑肿瘤的发病机制、新的治疗手段及精准治疗策略
近年以第一/通讯作者于iMeta(2023,IF=33.2)、Exploration(2025,IF=22.5,封面论文)、J Biomed Sci(2023;2024,IF=12.771)、J Big Data(2022;2023,IF=10.835)、Medcomm(2024,IF=10.7)等高水平期刊发表原创性论文23篇,参与专著出版一部
入选国家博士后海外引才计划、湖南省芙蓉计划科技创新类青年人才、中南大学湘雅医院飞凡人才计划
主持国家自然科学基金面上、青年项目,湖南省自然科学基金青年项目及中国博士后科学基金面上项目,累计在研科研经费200余万元
任iMeta及Exploration青年编委、中国免疫学会高级会员、多家高水平SCI期刊审稿人
助理研究员,医学博士,硕士研究生导师
以第一作者/通讯作者在Nucleic Acids Research(2020,IF=19.6)、Journal of Hematology & Oncology(2023,IF=28.5)、Journal of Biomedical Science(2024,IF=12.7)、Exploration(2025,IF=22.5,封面文章)、Cancer Research(2025,IF=16.6)等高水平杂志发表论文12篇
主持国家自然科学基金青年项目等课题5项;曾赴美国哈佛大学、匹兹堡大学进行联合培养;任iMeta及Exploration青年编委,多家高水平SCI期刊审稿人
教授、主任医师,医学博士,博士研究生导师
临床工作集中于颅底肿瘤的诊治,近10年累计主刀神经外科手术8000余例;在完成临床工作的同时,开展以临床问题为导向的基础研究,以第一作者或通讯作者在
Mol Cancer、Exploration、Science Advance、Neuro Oncol、J Big Data、Clin Transl Med、Mol Ther Nucleic Acids、iMeta、J Neurosurg、Neurosurgery等专业权威期刊发表学术论文50余篇,主编专业著作2部,主译3部
获评2024年湖南省科技创新领军人才,获教育部科技进步奖二等奖1项,获湖南省科技进步二等奖2项
主持国家重点研发计划、国家科技支撑计划项目、国家自然科学基金等国家和省部级项目10余项
担任中华医学会神经外科分会第七届青年委员会秘书长、湖南省医学会神经外科委员会副主任委员、中国脑膜瘤协作组副组长等学术职务
识别二维码,前往刘庆 教授学术主页
查看更多精彩内容
声明:脑医汇旗下神外资讯、神介资讯、脑医咨询、神内资讯所发表内容之知识产权为脑医汇及主办方、原作者等相关权利人所有。未经许可,禁止进行转载、摘编、复制、裁切、录制等。经许可授权使用,亦须注明来源。欢迎转发、分享。投稿/会议发布,请联系400-888-2526转3脑医汇App全新功能“AI问答”正式上线,您临床实践中的得力助手!欢迎点击“阅读原文”了解更多!